We got some amazing denovo genome assemblies of Pogonomyrmex barbatus! They were done as part of our collaboration with Deborah Gordon’s lab. These super-high-quality, near-chromosome-level assemblies will hopefully improve the performance of kinship inference that has been giving Felix a lot of trouble in this unorthodox population (dependent lineage system).
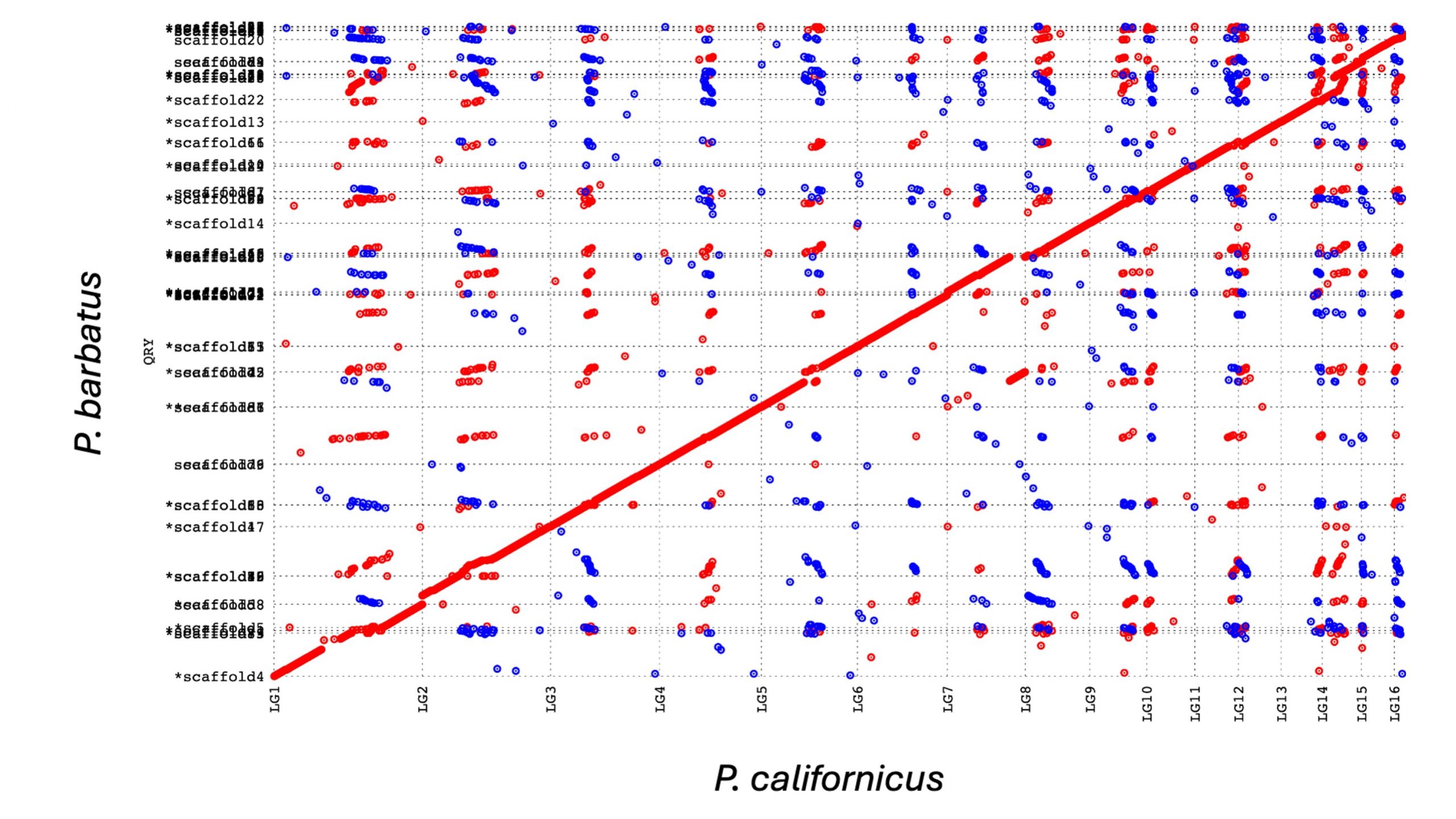
The assemblies were done in the Biotechnology Center at the University of Illinois. We just sent them the ants and they did everything. Huge shout out for Alvaro Hernandez and his team who got amazingly high quality data from a combination of PacBio and TellSeq from the DNA of a single ant, and also for Kim Walden who did an exceptionally thorough job with the assembly. I’ve never even heard about TellSeq before. The PacBio contigs were alright, but nothing special. It’s the TellSeq that got them nearly to chromosome level – scaffold N50 is over 10Mb! Check out this beautiful alignment to the (also exceptinally good) P. californicus genome. Kim even identified almost all of the chromosomes’ telomeres at the ends of most of the large scaffolds! Perhaps the coolest thing about all this is that Kim was actually part of the team that did the first P. barbatus genome that was published in 2011! One of the first 7 ant genomes that were published at the time.