Research
Multiple studies in the lab use population genomic and phylogenomic sequencing and computational molecular evolution analyses to scan genomes and search for the molecular basis of adaptations related to the social life of ants and other social insects, including: social communication based on chemical signals synthesis (e.g. synthesis of pheromones) and perceptions (e.g. olfactory receptors); variation of social behavior (e.g. foraging) and social structure (e.g. monogyne vs. polygyne). Along the way, we also contribute to the methods that others and we use, such as optimizing sequencing strategies for population genomics (see software).
Genetic basis for chemical communication in Cataglyphis desert ants
This project aims to identify genes responsible for chemical signaling in the Israeli ant Cataglyphis niger, using a combined approach of population genomic sequencing and chemical analysis. We started using this species as a model in collaboration with the lab of Abraham Hefetz in Tel Aviv University, who pioneered the study of this system using chemical ecology approaches. This species is also of interest because it displays a social polymorphism similarly to fire ants (see below).
 Tal and Daphna processing Cataglyphis ants collected from Bezet Beach
Tal and Daphna processing Cataglyphis ants collected from Bezet Beach
Before we can analyze the population genomics of this species, we first need a reference genome sequence. This was the MSc project of Tal: sequencing and assembling the reference genome for Cataglyphis niger. He also compared alternative approaches to de novo genome sequencing and gene annotation, in the special case of haplodiploid species and showed that it’s advantageous to use a single haploid male for both DNA and RNA extraction. Actually, the contig N50 size of the genome assembly made from a haploid male sample was three times bigger than the one made from his diploid sister!
The reference genome serves as the foundations for population genomic studies. First, we used 117 brothers from a single nest for QTL mapping study. Shani used these samples for both genomic sequencing and chemical analysis of cuticular hydrocarbons (CHC; the pheromones used by ants for nestmate recognition). She identified 31 different long-chain CHCs in variable amounts on different ants from different colonies. She then constructed the first RADseq library ever done in Israel (Restriction-associated DNA sequencing), and the results came out top quality:
These raw sequence reads then went through a lot of processing and bioinformatics, done by Pnina and Besan. Besan constructed a linkage map for the 26 chromosomes of C. niger, with the help of Abraham Korol and using his software MultiPoint. She then conducted QTL mapping of the chemical traits of these ants (quantities of cuticular hydrocarbons) using the software MultiQTL, also from the Korol lab. Almost every chromosome has some hint of QTLs. After thorough analysis and correction for multiple testing (FDR < 10%) we ended up with 20 QTL that had statistically significant association with one of nine CHCs. Most of these CHCs had more than one QTL (and up to four), suggesting that some CHCs have a stronger genetic basis than others.
GWAS: We also collected a much more extensive population sample, from 50 colonies in Betzet beach. Shani constructed whole-genome libraries of 6 workers from each colony (300 genomes altogether). Combined with chemical analysis of the same samples, these results allow us to test for an association between genetic variation in specific genes and variation in the cuticular hydrocarbons i.e. a genome-wide association study (GWAS), which is conducted by Pnina.
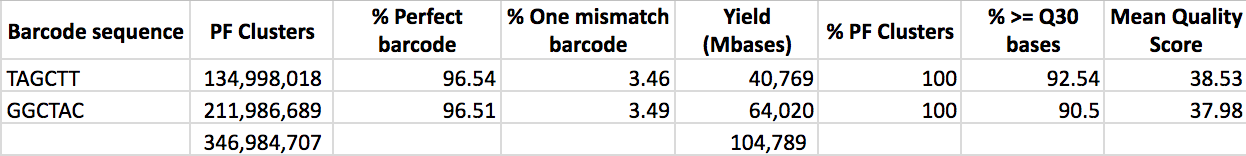
Social polymorhpism associated with a supergene in Cataglyphis niger: Previous studies identified three clades within this species complex: C. niger, C. savignyi, and C. drusus. The three were distinct monophyletic clades according to their mitochondrial haplotypes, and they also display distinct social colony structures: C. niger is polygyne, whereas C. savignyi and C. drusus are monogyne. In a joint paper with the Hefetz lab (Reiner-Brodetzki et al. 2019) we used RAD sequencing and population structure analysis of thousands of SNP loci to show that these three mitochondrial clades are not monophyletic in terms of their nuclear DNA. In fact, there is no detectable genetic divergence among them, and they form one un-differentiated population cluster in the STRUCTURE results:
STRUCTURE results showing that the niger, drusus, and savignyi mitotypes are actually one socially polymorphic species
We also conducted a maximum likelihood test for gene flow (using 3s) and rejected the null model of no gene flow between these clades. That is, there is significant evidence for gene flow between all these populations and between colonies of different social structure. Finally, we showed that the spermatheca of polygyne queens with the C. niger mitotype contains mostly sperm with the C. savignyi mitotype. That is, polygyne queens mate mostly with males from monogyne colonies. Therefore, C. niger is one species that harbors a social polymorphism. Naturally, our next goal is to reveal the molecular basis for this polymorphism. Is it genetic? epigenetic? Is it a supergene as in the case of other ants such as Solenopsis invicta and Formica selysi?
Aparna is leading a big pop-seq project dissecting the genomic basis of this polymorphism. We collected 30 whole nests from a polymorphic population (monogyne and polygyne), and conducted aggression assays to help characterize the social form (polygynes are typically less aggressive). Then Aparna did RAD-seq for >20 ants per nest. This was the largest sample size ever done in the lab, and our Robot Noah helped a bit (check out this video to see it in action). The analysis revealed a large supergene on one of the chromosomes, broadly similar to those previously described in the other species.

Aparna doing bioinformatics (analyzing RADseq data from our pilot) while monigoring Robot Noah remotely as it constructs 800 RAD libraries from monogyne and polygyne samples from the polygyne Cataglyphis niger population in Tel Baruch
Evolution of social structure in Solenopsis fire ants
Another study system in our lab is the fire ant Solenopsis invicta. This species is a useful model because the genetic basis for a Mendelian trait affecting social behavior is known to be a non-recombining “social chromosome” that is similar to a Y sex chromosome (Wang et al. 2013). This chromosome determine the behavior of ants in this species leading to dramatic differences in their social structure: one social form allow only one queen per nest (monogyne) while the other form accept many additional queens to the same nest (polygyne). The recent evolution of this system involved many micro- and macro-mutations that alter social behavior and organization. We use inter-species comparative genomics and intra-species population genomics to study the evolution of this social chromosome and other genetic elements that underlie the various biological functions responsible for social behavior, especially chemical communication.
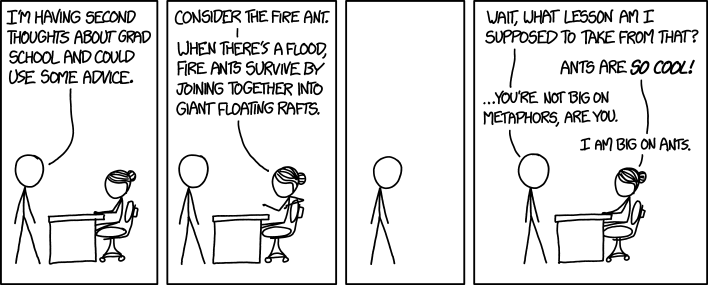
The population genetics of this species is also interesting as a case study of a high-impact invasive species. Fire ants were inadvertently introduced from north Argentina to Alabama, USA in the 1930’s. They quickly grew to very large and dense populations, spread across south-eastern USA and later were introduced from the US to various islands, Australia and China. This species causes great damages to both ecology and agriculture. Annual losses of $750 million were estimated in the United States alone.
In a collaboration with the lab of DeWayne Shoemaker (USDA) we use these well studied populations to investigate how a social insect becomes adapted to invasiveness. By sequencing whole genomes of many individuals from population samples we identify hundreds of thousands of polymorphic sites across the genome and compare their allele frequencies between native and introduced populations. In her PhD studies, Pnina applies a range of statistical inference methods to reconstruct the recent history of these populations and identify genomic regions that experienced positive selection. These genes may be responsible for recent local adaptation of different populations in the native and the introduced range.
Invasion history of the fire ant Solenopsis invicta.
Evolution of foraging strategy in varying climate
This project is a collaboration with the lab of Deborah Gordon (Stanford) funded by an NSF-BSF grant. Deborah’s lab has been studying variation in foraging strategies in desert haverster ants (Pogonomyrmex barbatus), which is the colonies response to variation in conditions (especially humidity). Felix is constructing genomic libraries for hundreds of colony samples and using this genomic dataset to infer kinship among colonies. Thereby we hope to be able to quantify reproductive success at the colony level, and test whether it is associated with the variation in foraging.
Universal adaptations of ants – olfactory receptors
The first whole genome sequencing of seven ant species allowed us to search for genes and gene families underlying adaptations of ants in general. Whole genomes scans for positive selection using dN/dS tests (site and branch-site tests in PAML) identified genes implicated in functional categories related to morphology, metabolism, and behavior. Focused analyses of odorat receptors (ORs) from two ant species (about 300 per species) identified subfamilies of ORs that underwent rampant ant-specific gene duplications and positive selection. These analyses generated candidate genes for further investigation of adaptive evolution of communication in ant societies.
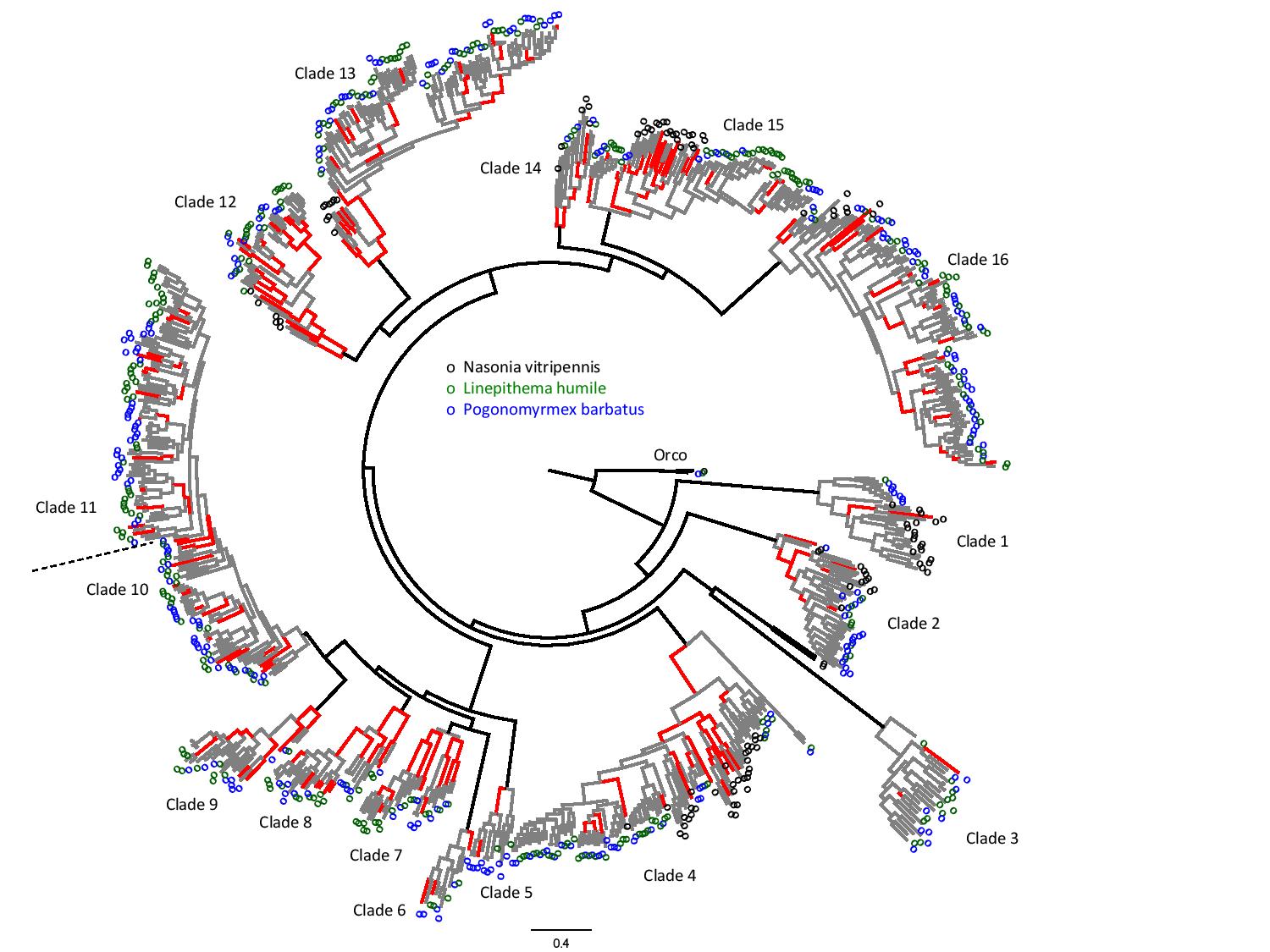
Gene tree of odorant receptors from two ant species (green and blue) and one wasp (black). Branches with positive selection are marked in red (Roux et al. 2014).
In her MSc thesis, Rana expanded this analysis to many more ant species, including Solenopsis fire ants (Saad et al. 2018). Amir conducted a focused study of a subfamily of ORs found on the fire ant social chromosome (Cohanim et al. 2018). These analyses revealed specific receptors that underwent dramatic evolution in the fire ant lineage.
More recently, as part of the GAGA Consortium (Global Ant Genomics Alliance), we are working on a much larger dataset of ORs annotated for high-quality genomes of 163 ant species (hundreds of ORs per species). Yoann is searching for an association between the evolution of social complexity and adaptive evolution of ORs.
Evolution of novel odorant specificities in odorant receptors
This project is a collaboration with the labs of Juergen Liebig (ASU) and Mickey Kosloff (U Haifa) funded by an NSF-BSF grant. We’re looking at ORs that underwent recent adaptive evolution in Camponotus floridanus (this part is done by Yoann from our lab) to identify amino acid substitutions in the receptor’s ligand binding pocket that altered its specificity towards novel cuticular hydrocarons (CHCs).
Evolution of sex determination in ants and bees
All 200,000 Hymenoptera species (including ants, bees, wasps and sawflies) have haplodiploid sex determination. They do not have sex chromosomes. Instead these insects may be either haploids that develop into males, or diploids that develop into females. While this general principle is common to all species in this large insect order, the underlying molecular genetic mechanisms are diverse and dynamic throughout evolution. One type of mechanism is complementary sex determination (CSD). CSD was first discovered in the honey bee Apis mellifera. Also the genes responsible for this mechanism were first discovered in the honey bee: feminizer and csd. These two genes are homologs of the transformer gene found in other insects. They were thought to be the result of a gene duplication event in the honey bees.
In one of the first studies of sex determination genes in ant genomes we discovered that almost every ant genome sequenced has two copies of this gene, like the honey bee. Surprisingly, each ant lineage appears to have its own independent duplication of the ancestral gene transformer. We put forward the hypothesis that this pattern is the result of a single ancestral duplication followed by concerted evolution of the two copies (Privman et al. 2013).
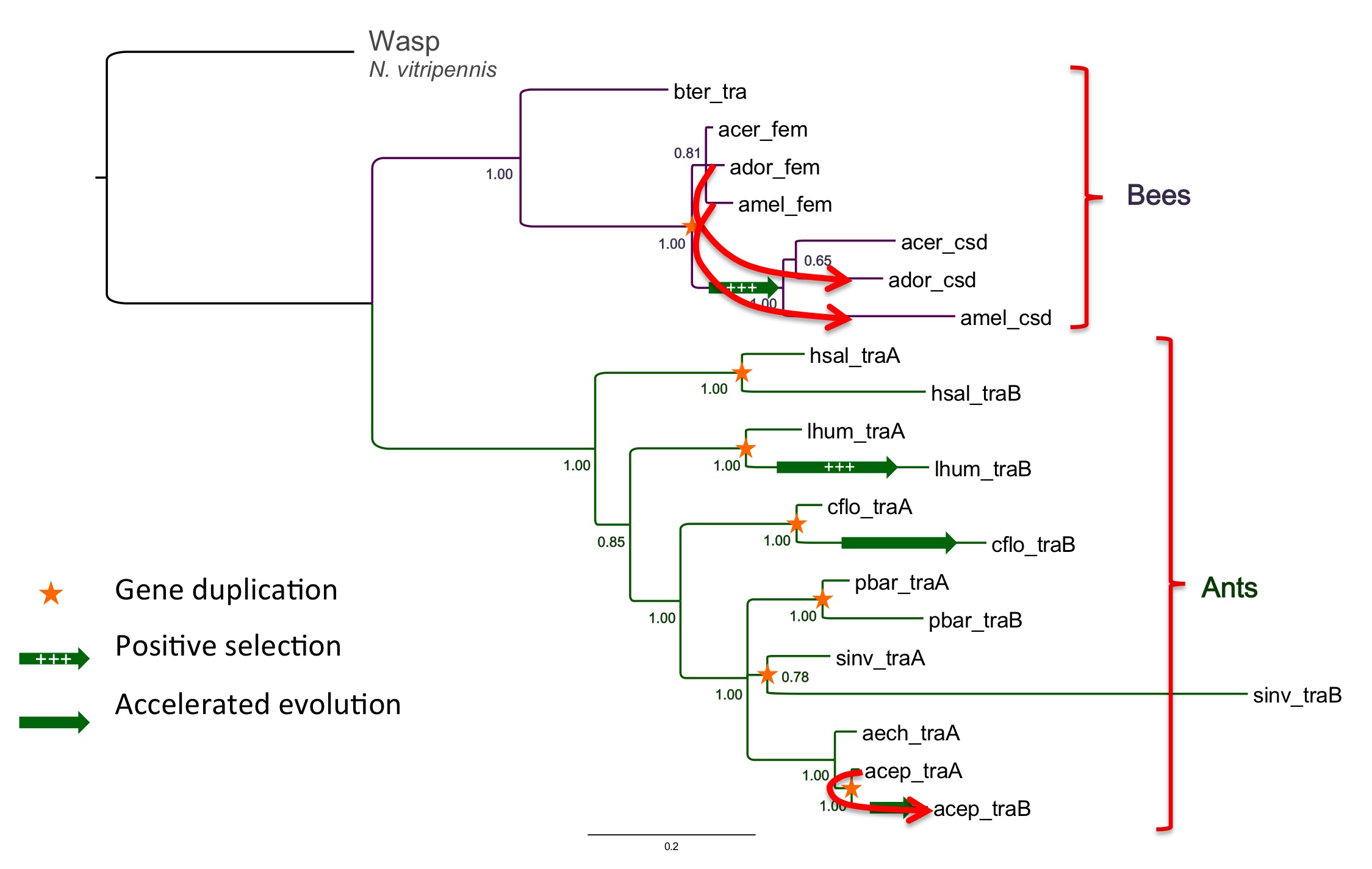
Gene tree of transformer homologs in ants and bees. Stars mark apparent gene duplications. Arrows indicate evidence for concerted evolution (gene conversion events)
More recently, we are collaborating with the lab of John Wang to characterize the sex determination locus of the fire ant Solenopsis invicta and its population genetics.

